Review Article
Staphylococcus aureus Alters Host Epigenetics for Immune Evasion: Insights
- Dr. Swarup K. Chakrabarti
Corresponding author: Dr. Swarup K. Chakrabarti, H. P. Ghosh Research Center, HIDCO (II), EK Tower, New Town, Kolkata, West Bengal 700161, India.
Volume: 2
Issue: 5
Article Information
Article Type : Review Article
Citation : Swarup K. Chakrabarti, Dhrubajyoti Chattopadhyay. Staphylococcus Aureus Alters Host Epigenetics for Immune Evasion: Insights. Journal of Medical and Clinical Case Reports 2(5). https://doi.org/10.61615/JMCCR/2025/OCT027141009
Copyright: © 2025 Dr. Swarup K. Chakrabarti. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
DOI: https://doi.org/10.61615/JMCCR/2025/OCT027141009
Publication History
Received Date
19 Sep ,2025
Accepted Date
04 Oct ,2025
Published Date
09 Oct ,2025
Abstract
Staphylococcus aureus (S. aureus) is a formidable pathogen in humans that can lead to a diverse range of infections, from mild skin issues to serious systemic illnesses. Its frequent appearances and endurance are often linked to its ability to resist both the host's immune responses and antibiotic treatment. Recent research sheds light on a new and previously unexplored aspect of its pathogenesis—its ability to affect the host's epigenetic mechanisms. This examination investigates how S. aureus can modify host chromatin structure through changes in DNA methylation, alterations in histone proteins, and influences on non-coding RNAs, ultimately resulting in the reprogramming of immune responses and cellular processes that support bacterial survival and longevity. This article elaborates on the ways S. aureus affects host cell functions, pointing out potential pathways for enhancing infection biology, diagnostics, and targeted treatments. Consequently, this article encourages further exploration of the epigenetic interactions between host and pathogen as a vital area in combating bacterial persistence.
Keywords: Staphylococcus aureus, Antibiotic resistance, Epitherapeutics, Virulence regulation, Epigenetics, Microbial pathogenesis
►Staphylococcus aureus Alters Host Epigenetics for Immune Evasion: Insights
Swarup K. Chakrabarti1*, Dhrubajyoti Chattopadhyay1, 2
1H. P. Ghosh Research Center, New Town, Kolkata, West Bengal 700161, India.
2Sister Nivedita University, New Town, West Bengal 700156, India.
Introduction
Staphylococcus aureus (S. aureus) is a spherical, Gram-positive bacterium that belongs to the Micrococcaceae family. It is catalase-positive and oxidase-negative—two key features commonly used for its identification in clinical microbiology [1,2]. Notably robust and versatile, S. aureus frequently exists as an asymptomatic member of the human microbiome, particularly on the skin and within the anterior nares (nasal cavities). For most individuals, this colonization is harmless. However, S. aureus can change from a harmless commensal to a powerful opportunistic pathogen in situations where the immune system is compromised, such as skin damage or systemic immune suppression [3,4]. Once it crosses the host’s protective barriers, it can lead to a variety of health issues—ranging from mild skin infections such as impetigo, boils, and carbuncles to more serious and potentially fatal conditions like necrotizing pneumonia, sepsis, osteomyelitis, infective endocarditis, and toxic shock syndrome [5,6].
This diverse pathogenic potential is driven by a sophisticated arsenal of virulence factors, including surface adhesins, tissue-degrading enzymes, and powerful exotoxins [7]. These factors are tightly regulated by global regulatory systems, most notably the accessory gene regulator (agr) and staphylococcal accessory regulator (sar) operons, which modulate virulence gene expression in response to environmental cues [8,9]. In addition to invasive disease, S. aureus is also a significant driver of foodborne illness. Enterotoxigenic strains can contaminate improperly handled or stored foods, resulting in staphylococcal food poisoning—characterized by the abrupt onset of vomiting, nausea, and abdominal pain. The thermostability of these enterotoxins allows them to remain biologically active even after exposure to regular cooking temperatures, thereby posing a persistent public health concern [10,11]. Together, the combination of S. aureus’s virulence, adaptability, and growing antibiotic resistance renders it a constantly evolving threat to human health. A deeper understanding of its pathogenic mechanisms and regulatory systems is crucial for the development of effective diagnostics, preventive strategies, and therapeutic interventions aimed at mitigating its global impact.
On the other hand, epigenetics, the study of inheritable changes in gene expression that do not involve alterations to the DNA sequence itself, has emerged as a crucial field for understanding how host-pathogen interactions unfold at the molecular level. These epigenetic mechanisms—comprising DNA methylation, histone modifications, and non-coding RNA interactions—play essential roles in regulating biological processes such as cell differentiation, development, and responses to environmental signals [12,13]. Traditionally viewed as tools of cellular memory and gene control, epigenetic modifications are now recognized as pivotal players in infectious disease progression. Specifically, S. aureus has evolved complex strategies to alter the host's epigenetic landscape in ways that benefit its own survival and proliferation. By modifying the host’s epigenetic landscape, S. aureus can impair immune defenses, subvert host cellular functions, and promote its own survival—even in the presence of antibiotic treatment [14,15]. These manipulations allow S. aureus to evade immune surveillance and create a supportive environment within the host. While genetic mutations are well-known contributors to antibiotic resistance, epigenetic changes add a new dimension to this challenge. S. aureus can temporarily enhance its resistance by altering host immune signaling pathways or suppressing antimicrobial peptide production [16,17]. This not only helps the pathogen persist under adverse conditions but also complicates treatment outcomes. In addition, S. aureus engages directly with the host’s chromatin regulatory systems. Through secreted bacterial factors, it can induce histone modifications and alter DNA methylation patterns in host cells. These changes disrupt normal host cell functions—including immune responses, cell cycle regulation, and apoptosis—thus creating an environment that favors bacterial reproduction and long-term persistence [18,19]. Such host-pathogen epigenetic interactions reveal a dynamic and intricate relationship in which S. aureus actively reprograms the host’s molecular environment for its benefit.
Hence, this review seeks to explore the ways in which S. aureus affects host epigenetic mechanisms to evade immune responses and the consequences, including antibiotic resistance. By exploring its effects on DNA methylation, histone modification, and other epigenetic alterations in host cells, the article aims to investigate the molecular mechanisms that S. aureus uses to evade the immune response and persist during antibiotic therapy. A comprehensive understanding of these mechanisms can illuminate the complex interplay between infection and immunity and highlight promising new therapeutic strategies that target both microbial and host epigenetic systems. In doing so, this review underscores the profound influence of epigenetic interactions on infection outcomes and their potential as leverage points in combating chronic and resistant S. aureus infections.
Structure of the Review Article
This review is organized into interrelated sections. We begin by examining the key S. aureus regulatory systems (agr and sar) and their influence on host epigenetic modulation. We then explore the molecular mechanisms by which S. aureus reprograms the host epigenome to evade immune detection and clearance. Subsequent sections address the potential involvement of host epigenetic pathways in S. aureus antibiotic resistance, highlighting how these interactions may compromise therapeutic efficacy. Finally, we synthesize current knowledge to assess the broader implications for understanding host–pathogen interactions and guiding future therapeutic strategies.
agr and sar: Key Regulators in S. aureus Virulence
The agr system of S. aureus is a core quorum-sensing mechanism that controls virulence gene expression in response to changes in bacterial population density. It operates by sensing autoinducing peptides (AIPs) secreted into the environment, which initiate a two-component signal transduction cascade. This cascade ultimately activates RNAIII, a regulatory RNA that coordinates the timing of virulence factor expression [20]. Early in infection, when cell density is low, agr activity remains subdued, allowing for the expression of surface-associated adhesins that promote host colonization and biofilm (organized bacterial communities covered with a matrix that they have created themselves) formation [21,22]. As bacterial numbers increase, enhanced agr signaling shifts gene expression toward the production of secreted toxins and enzymes, which facilitate tissue invasion and immune evasion [23]. Closely linked to agr is sarA (staphylococcal accessory regulator A), a global transcriptional regulator that influences the expression of both surface proteins and extracellular factors. The sarA gene is transcribed through three overlapping promoters—P1, P2, and P3— located upstream of the coding sequence. These promoters generate multiple transcription start sites, leading to different lengths of transcripts that encode a single SarA protein [24]. While P1 and P2 are consistently active or weakly controlled under baseline conditions, P3 serves as the most efficient and precisely regulated promoter, responding dynamically to environmental stimuli and control signals. The SarA protein functions by directly binding to the promoter regions of its target genes or by modulating the activity of other transcriptional regulators [25]. This enables S. aureus to finely tune its virulence arsenal in response to environmental cues. Additionally, sarA regulates agr activity directly, forming an intricate regulatory feedback loop that dynamically governs adhesion, invasion, and immune subversion depending on the infection stage and host immune response [26]. Moreover, S. aureus adds an extra layer of regulation by using SigB, an alternative sigma factor—a protein that assists in initiating the transcription of specific gene groups [27]. When triggered by environmental stresses such as heat shock, osmotic stress, or nutrient limitation, SigB binds to the core RNA polymerase to form a holoenzyme complex. This shift enables the bacterium to prioritize transcription of stress-response genes, allowing rapid adaptation to adverse conditions [28]. SigB is encoded within a gene cluster that includes several anti-sigma factor genes, ensuring precise control of its activity. While primarily associated with stress adaptation, SigB also broadly influences virulence gene expression [29]. By interacting with both agr and sarA, it facilitates the transition from acute to chronic infection, enhancing survival within the host and resistance to immune clearance [30].
Collectively, these regulatory systems orchestrate the expression of a wide array of virulence factors. These include alpha-hemolysin (Hla), a pore-forming toxin that targets epithelial and immune cells; phenol-soluble modulins (PSMs), which contribute to inflammation, cell lysis, and biofilm architecture; and Protein A (SpA), a surface protein that binds to the Fc region of immunoglobulins, thereby inhibiting opsonization (the process by which pathogens are marked for ingestion) and phagocytosis [31,32]. The agr/sarA regulatory axis also controls the expression of superantigenic toxins such as enterotoxins and toxic shock syndrome toxin-1 (TSST-1), which hyperstimulate T cells and can lead to cytokine storms, septic shock, and multi-organ failure [33,34]. Additionally, numerous adhesins and hydrolytic enzymes—including proteases and lipases—aid in tissue degradation, immune evasion, and nutrient acquisition. Emerging findings also suggest that many of these virulence factors function not only as direct mediators of pathogenesis but also as modulators of host immune and epigenetic responses. For example, Hla and PSMs can influence host cell signaling and the expression of inflammatory genes by altering chromatin accessibility, histone modifications, and DNA methylation [35,36]. These epigenetic changes can modulate immune responses and contribute to immune tolerance, persistent infection, or lasting post-infectious conditions.
In summary, the agr, sarA, and SigB regulatory systems are central orchestrators of S. aureus pathogenesis, enabling the bacterium to dynamically adapt to host defenses and persist through various phases of infection. Understanding these regulatory mechanisms and their downstream effects enhances our comprehension of bacterial virulence and opens new avenues for anti-virulence therapeutic strategies. By targeting these key regulators rather than essential growth processes, it may be possible to neutralize the pathogen while reducing the selective pressure that drives antibiotic resistance—offering a promising direction for future treatments.
How agr/sar-Regulated Virulence Factors Modulate Host Epigenetics
Histone Modifications
As previously mentioned, S. aureus employs a sophisticated network of virulence regulators, agr and sar systems, to control the expression of factors that influence host-pathogen interactions. Beyond their cytotoxic and immune evasion roles, recent findings indicate that several of these virulence factors can alter host epigenetic landscapes. By managing chromatin structure and transcriptional properties, these elements enable the pathogen to alter host cell responses, enhancing inflammation, promoting immune suppression, and aiding bacterial persistence. The following section explores the impact of centrally agr/sar-regulated toxins, such as Hla and PSMs, on host epigenetic processes.
S. aureus Hla forms heptameric transmembrane pores in the membranes of host cells, causing ion dysregulation, especially a rapid influx of calcium ions. In human airway epithelial cells, the influx of calcium initiates intracellular signal transduction pathways, like the p38 MAP (Mitogen-Activated Protein Kinase) pathway, which mediate inflammatory responses [37,38]. Hla prompts both murine and human monocytic cells to trigger the NLRP3 (NOD-, LRR-, and Pyrin domain-containing protein 3) inflammasome, resulting in the release of pro-inflammatory cytokines and cell death [39]. Although the direct connections between calcium influx and chromatin remodeling are yet to be established, downstream signaling pathways suggest a role in gene expression and epigenetic regulation. For instance, calcium/calmodulin-dependent kinases (CaMKs) that are triggered by increased calcium concentrations can phosphorylate and stimulate CBP/p300 (CREB-Binding Protein), significant histone acetyltransferases (HATs), leading to the acetylation of histones H3 and H4 and the transcriptional activation of inflammatory genes such as IL (interleukin)-8, TNF-α (Tumor Necrosis Factor-alpha), and CXCL1 (C-X-C motif chemokine ligand 1) [40]. In accordance, Hla has been shown to cause reversible changes in the accessibility of chromatin within endothelial and epithelial cells, demonstrating a dynamic and toxin-responsive epigenome [41]. In addition to Hla, PSMs—small amphipathic peptides regulated by the agr quorum-sensing system—also modulate host responses by eliminating neutrophils and monocytes, leading to pyroptosis and necroptosis [42]. Conversely, PSMs enhance the trimethylation of histone H3 at lysine 4 (H3K4me3), a chromatin modification associated with activation, particularly on cytokine promoters induced by NF-κB (Nuclear Factor kappa-light-chain-enhancer of activated B cells) such as IL-1β, IL-6, and CCL20 (C-C motif chemokine ligand 20/Macrophage Inflammatory Protein 3-alpha) [43-45]. They also enhance the secretion of high mobility group box 1 (HMGB1) into the extracellular environment, a nuclear protein that functions as an alarmin, capable of triggering chromatin remodeling and DNA demethylation upon TLR4 (Toll-Like Receptor 4) activation [46,47]. These virulence factors together orchestrate complex epigenetic reprogramming of host cells, shaping inflammatory and immune responses to S. aureus infection [48,49].
DNA Methylation
Chronic infections and inflammation due to S. aureus are often triggered by agr/sar-regulated toxins that stimulate immune activation while also leading to significant epigenetic changes in the immune cells of the host. These toxins generate reactive oxygen species (ROS) and elevate pro-inflammatory cytokines, creating a microenvironment conducive to epigenetic reprogramming [50,51]. In persistent skin infections, dermal macrophages exhibit hypomethylation at the promoters of inflammatory genes such as IL-6, TNF-α, and TLR2. This is likely facilitated by ROS-induced suppression of DNA methyltransferase 1 (DNMT1), leading to chronic and possibly unregulated inflammatory responses [52,53]. Similarly, in macrophages derived from human monocytes, exposure to supernatants from agr-positive S. aureus leads to the downregulation of DNMT3b, which further amplifies the expression of inflammatory genes [54,55]. These findings emphasize the pathogen's capability to alter host DNA methylation processes to establish an inflammatory setting favorable for its persistence. Adding to this complexity, S. aureus additionally releases superantigens like TSST-1, the production of which is also regulated by the agr and sar systems. TSST-1 serves as a potent activator of CD (Cluster of Differentiation) 4+ T cells, resulting in widespread DNA hypomethylation and targeted demethylation at the IFN-γ (Interferon-gamma) promoter, thereby steering the immune response towards a pro-inflammatory Th1 phenotype [56,57]. TSST-1 can potentially disrupt the epigenetic terrain and epigenetic profiling of regulatory T cells (Tregs), particularly affecting the DNA methylation patterns at the Forkhead box P3 (FoxP3) gene locus [58]. These epigenetic changes impede the stable expression of FoxP3, a crucial transcription factor necessary for Treg lineage determination and suppressive function, thus undermining their immunoregulatory identity and performance. Since proper DNA methylation is essential for the stability of the Treg lineage, this epigenetic disruption may lead to systemic inflammation and immune imbalance during S. aureus infections. These combined mechanisms illustrate how S. aureus skillfully exploits host epigenetic processes to influence immune responses and sustain chronic infection [59].
Non-coding RNAs
RNAIII acts as the main effector within the agr system of S. aureus, produced from the P3 promoter. In addition to the P3 promoter, the agr operon has a P2 promoter, which drives the expression of the quorum-sensing components (agrBDCA) [60,61]. In this system, agrB encodes a membrane-bound protease that processes AgrD into the autoinducing peptide (AIP); agrD encodes the AIP precursor; agrC encodes a membrane-bound sensor kinase that detects AIP; and agrA encodes the response regulator that activates transcription of target genes, including RNAIII. To further elaborate, when AgrA is activated, it boosts the activity of both promoters, creating a feedback loop. RNAIII then regulates the expression of virulence genes after transcription, influenced by cell density and environmental cues; this regulation is also observed during periods of low bacterial cell density [62,63]. These include exoproteins and surface-associated proteins that contribute to immune evasion, tissue damage, and inflammation. Although RNAIII functions inside the bacterium, its downstream targets—many regulated alongside the sar system—significantly influence host immune signaling. A notable area of research involves the ability of agr-active strains to influence host non-coding RNA networks, particularly those involved in immune regulation and inflammation [64,65].
MicroRNAs (miRNAs) play a crucial role as post-transcriptional regulators of gene expression and have shown a dynamic response to bacterial infection. In S. aureus infection, miR-146a and miR-155 are consistently upregulated in monocytes and dendritic cells, mainly because of TLR signaling, a route usually triggered by bacterial elements [66,67]. These miRNAs precisely adjust inflammation by targeting negative regulators of the NF-κB pathway, such as IRAK1 (Interleukin-1 Receptor-Associated Kinase 1) and TRAF6 (TNF Receptor-Associated Factor 6) [68,69]. The upregulation of these miRNAs enhances NF-κB activity, leading to the ongoing generation of pro-inflammatory cytokines. Furthermore, they can initiate a feed-forward loop of inflammation by promoting the epigenetic silencing of anti-inflammatory feedback systems, which subsequently enhances the host immune response. While direct evidence linking RNAIII to alterations in the host miRNA profile remains limited, studies suggest that virulence factors controlled by agr may indirectly affect the host’s miRNA landscape [70,71]. Notably, infections from agr mutants have shown a connection with diminished miR-146a and miR-155 responses, indicating that virulence factors regulated by quorum sensing are necessary to activate these miRNA-dependent inflammatory pathways [72,73]. This discovery supports the idea that S. aureus can influence post-transcriptional gene expression in the host through miRNA pathways. In addition to miRNAs, long non-coding RNAs (lncRNAs) also play a role in regulating host responses to S. aureus. LncRNAs such as lnc-IL7R (Long Non-Coding Interleukin 7 Receptor) and MALAT1 (Metastasis-Associated Lung Adenocarcinoma Transcript 1) are expressed at varying levels during infection and have demonstrated interactions with chromatin modifiers, transcription factors, and enzymes that modify histones [74-76]. For example, MALAT1 has been discovered to play a role in regulating the status of histone methylation and acetylation, thereby influencing the transcription of genes related to the immune system [77]. By modifying the epigenetic environment, these lncRNAs can enhance the expression of inflammatory genes initiated by bacterial infection, thereby increasing the duration and severity of the immune response.
In summary, S. aureus exploits the host's non-coding RNA systems through agr/sar-regulated virulence factors to influence immune mechanisms at both the epigenetic and transcriptional levels. Although the direct impacts of RNAIII on host non-coding RNAs (ncRNAs) require more extensive investigation, current data emphasizes intricate interactions between bacterial signaling networks and host epigenetic regulators. This communication across species highlights the potential of focusing on non-coding RNA pathways as part of treatment strategies to combat inflammation and disease in S. aureus infections.
- Escape of S. aureus Through Host Epigenome Modulation
S. aureus employs sophisticated strategies to evade the host’s immune system by changing its gene expression through epigenetic modifications [78,79]. These adjustments can both trigger immune responses and suppress them, allowing the bacteria to survive longer within the host. This dual approach—either activating or weakening the immune defense—depends heavily on the timing, the amount of exposure, and the length of contact with the bacteria’s harmful factors, especially those regulated by the agr and sar systems. By shifting between promoting inflammation and dampening it, S. aureus can avoid being cleared by immune defenses, enabling it to stay in the host for an extended period. This ability allows the bacteria to cause both acute infections and establish persistent, long-term colonization. Interestingly, after initial contact with S. aureus or its secreted products, innate immune cells such as monocytes and macrophages undergo epigenetic reprogramming, equipping them for a more robust response upon subsequent exposure. This leads to what is referred to as trained immunity through the accumulation of activating histone marks—H3K4me3 and H3K27ac—at the promoters and enhancers of the key pro-inflammatory genes TNF-α, IL-6, and IL-1β [80- 82]. These chromatin alterations boost transcriptional readiness, even when triggered by unrelated pathogens or pathogen-associated molecular patterns (PAMPs) [83]. Moreover, a central element of the epigenetic memory caused by S. aureus is its metabolic foundation. In activated macrophages, exposure to pathogens triggers a shift to aerobic glycolysis, facilitated by the Akt (Protein Kinase B)-mTOR (mechanistic Target of Rapamycin)-HIF-1α (Hypoxia-Inducible Factor 1-alpha) signaling pathway [84,85]. The activation of HIF-1α enhances the expression of glycolytic transporters and enzymes, promoting metabolic reprogramming, while the buildup of metabolites such as succinate further stabilizes HIF-1α and boosts its activity [86]. Besides regulating metabolism, HIF-1α also recruits HATs to the promoters of inflammatory genes, resulting in chromatin alterations that enhance transcriptional accessibility and sustain the expression of pro-inflammatory genes [87]. Notably, these epigenetic alterations persist for weeks, rendering immune cells responsive to later inflammatory triggers well after the initial contact with S. aureus. This enduring alteration of transcription suggests that the host epigenome acts as a molecular memory of past microbial encounters, potentially leading to chronic inflammation after repeated or unresolved infections. Simultaneously, it is noted that trained immunity in macrophages induced by S. aureus can enhance protection against reinfection, emphasizing its dual role in host defense and disease development [88,89].
On the other hand, prolonged or cumulative exposure to S. aureus virulence factors may result in a condition of immune tolerance, characterized by a reduced responsiveness of innate immune cells [90,91]. This tolerized state is associated with the addition of repressive histone modifications, such as H3K9me2/3 and H3K27me3, at the promoters of inflammatory genes, alongside DNA hypermethylation in the regulatory regions of genes, including TNF-α, IL-1β, and CXCL10 (C-X-C Motif Chemokine Ligand 10) [92,93]. These epigenetic changes prevent transcription and reduce the production of pro-inflammatory cytokines. Experimental models have demonstrated that prolonged stimulation with S. aureus leads to reduced cytokine production and the formation of these suppressive chromatin modifications. Clinically, monocytes derived from patients with S. aureus bacteremia have shown altered expression profiles indicative of a tolerant condition, although some studies suggest preserved pro-inflammatory responsiveness following ex vivo stimulation [94]. The histone methyltransferases (HMTs) EZH2 (Enhancer of Zeste Homolog 2) and G9a/EHMT2 (Euchromatic Histone Lysine Methyltransferase 2) play a crucial role in the formation and preservation of this tolerant phenotype [95]. Their inhibition has been shown to rejuvenate immune function and reverse tolerance, suggesting they might act as therapeutic targets. The relationship between trained immunity and immune tolerance induced by S. aureus relies on factors like dose, timing, and length of exposure to its virulence factors [96]. This dual role allows the pathogen to evade immune clearance during acute infection and achieve lasting persistence within the host.
The Potential Role of Host Epigenetics in S. aureus Antibiotic Resistance
Evaluating the impact of the host epigenome on the expression of antibiotic resistance genes (ARGs) in S. aureus, such as mecA (which confers resistance to methicillin), vanA and vanB (responsible for vancomycin resistance), ermA (conferring resistance to macrolides), ermB (resistance to lincosamides), ermC (resistance to streptogramin B), and others, is a crucial focus in infectious disease research [97]. This might clarify why certain individuals are more susceptible to S. aureus, including resistant strains like MRSA (methicillin-resistant Staphylococcus aureus), compared to others. Historically, bacterial resistance has been attributed to inherent microbial mechanisms, such as mutations and horizontal gene transfer (HGT), which involve the spread of resistance genes via mobile genetic elements like plasmids, transposons, and bacteriophages [98,99]. These components can swiftly move between bacteria, disseminating resistance characteristics. Nonetheless, emerging evidence suggests that the host epigenome may also play a crucial role in influencing pathogen behavior. The host's epigenome regulates crucial immune functions, such as inflammatory responses and the production of antimicrobial peptides, which influence the tissue environment that S. aureus encounters [100,101]. Suppression of immune defense via epigenetic processes could prolong bacterial survival, inducing conditions that encourage the activation or selection of ARGs [102,103]. Hence, the link between host epigenetics and bacterial resistance reveals a novel regulatory layer that may influence the severity of S. aureus infections. In this regard, Figure 1 illustrates the complex epigenetic interactions between S. aureus and its host. S. aureus manipulates the host's epigenetic mechanisms to hinder immune responses while simultaneously promoting the expression of its own ARGs. This allows the bacteria to endure the effects of antimicrobials and may somehow strengthen the epigenetic alterations in the host, thereby establishing a vicious cycle that aids immune evasion and exacerbates the severity of the infection.
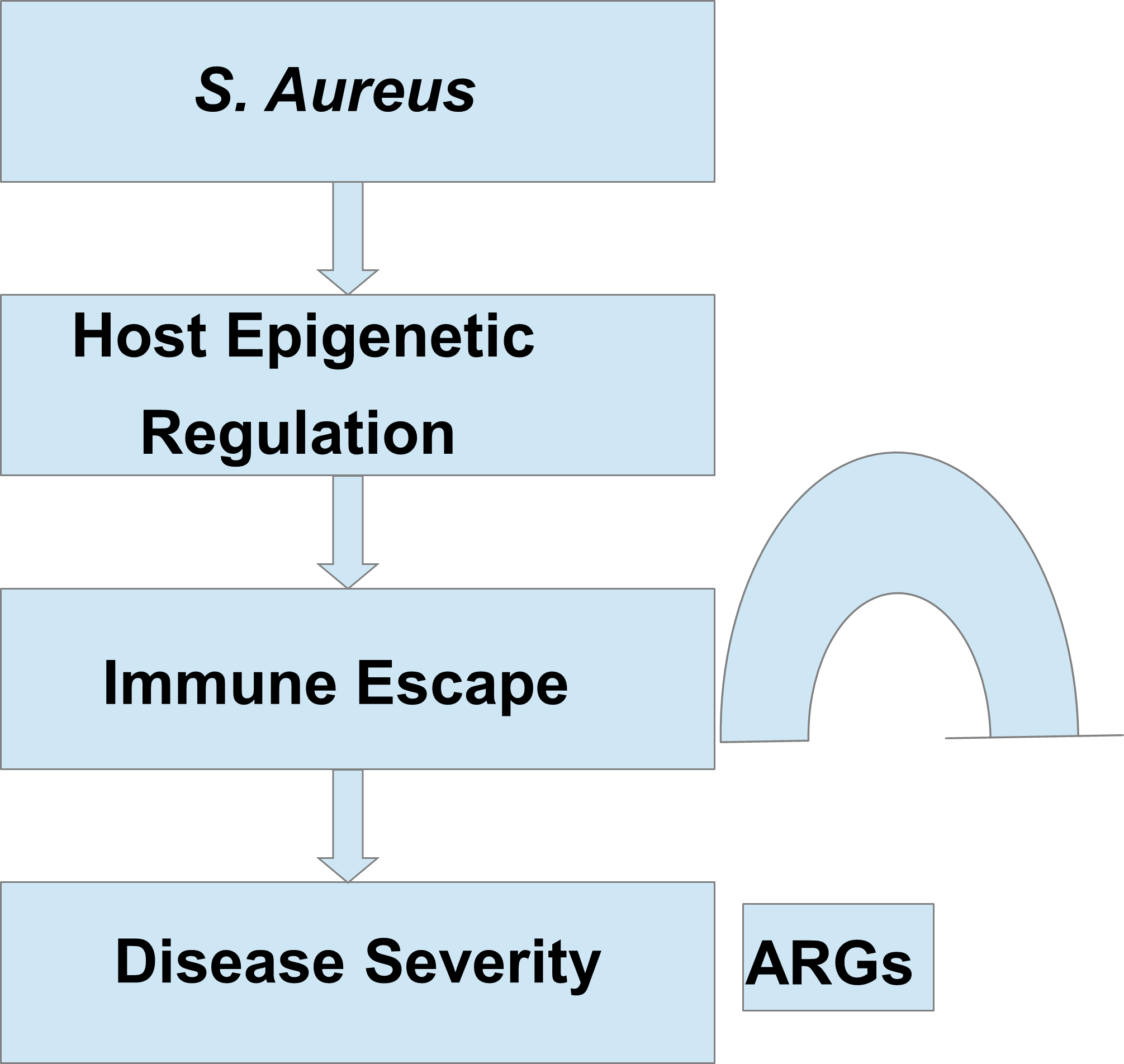
Legend 1. Epigenetic Interactions Between Staphylococcus aureus (S. aureus) and the Host. This diagram illustrates how S. aureus cleverly influences host epigenetic mechanisms to reduce immune system responses. At the same time, the bacterium increases the expression of antibiotic resistance genes (ARGs) to improve its chances of survival in the presence of antimicrobials. The heightened expression of ARGs may further affect the host's epigenetic regulation, creating a self-perpetuating feedback mechanism that fosters immune evasion and intensifies the severity of the infection.
Importantly, recurrent or chronic S. aureus infections are frequently observed in individuals with epigenetic dysregulations, such as those resulting from chronic inflammation, aging, or immune system suppression [104]. For example, aging is marked by a chronic low-level inflammatory state often referred to as "inflammaging." This condition results in ongoing activation of immune pathways, undermining immune defenses and increasing the likelihood of infections like S. aureus that exploit these imbalanced systems [105]. Additionally, chronic diseases related to aging, such as diabetes or vascular conditions, contribute to impaired wound healing, immune system dysfunction, and skin colonization, which increases the likelihood of S. aureus infection [106,107]. In these patients, epigenetic changes might result in weakened immunity, which can cause greater difficulty in combating infections and establish a cycle of recurrent attacks. Importantly, the host's epigenetic profile might influence the immune system's effectiveness against S. aureus, highlighted in the previous sections of this article, and its resistance strategies. For example, the methylation of DNA and histone modifications on immune-related genes could diminish the initial immune response, enabling S. aureus to survive longer and potentially gain the opportunity to develop resistance mutations or activate ARGs located on mobile genetic elements [108,109].
Moreover, epigenetic changes in the host triggered by S. aureus may form a feedback loop that supports the pathogen’s ability to maintain ARGs expression. This interaction between host and pathogen allows resistant strains to endure even when exposed to sublethal levels of antibiotics. The reprogramming of the host immune system at an epigenetic level can likewise contribute to the bacterium's increased capacity to avoid immune detection. For example, hypermethylation of gene promoter regions that code for pattern recognition receptors (PRRs) such as TLR2 and NOD2 (Nucleotide-binding Oligomerization Domain-containing Protein 2) or pro-inflammatory cytokines such as IL-6 and TNF-α can disrupt immune activation, allowing S. aureus colonization and immune evasion [110, 111]. This immunological tolerance would allow S. aureus to survive and flourish, particularly in situations of immune suppression or chronic inflammation, and boost the genetic expression of ARGs. Additionally, the epigenetic landscape of the host influences the microenvironment—not just local inflammation, oxygen levels, and nutrient availability—which in turn impacts the regulation of ARGs, including the expression of mecA related to methicillin resistance [112,113]. Moreover, local immune dysfunction and persistent inflammation can facilitate the formation of bacterial entities like biofilms or small-colony variants (SCVs, slow-growing bacterial subpopulations with altered metabolism), both associated with antibiotic resistance and long-term infections [114,115]. These adaptations are often triggered or sustained by the host's epigenetic state, which affects S. aureus behavior, virulence, and the acquisition of antibiotic resistance. In this context, as illustrated in Figure 2, the host's epigenetic processes—such as DNA methylation, modifications of histones, and alterations in chromatin structure—can influence the expression and transfer of microbial ARGs like mecA, vanA, vanB, ermA, and ermB. These relationships suggest that the host's epigenetic regulation of its microbiome could play a crucial role in shaping antibiotic resistance dynamics and the efficacy of treatment.
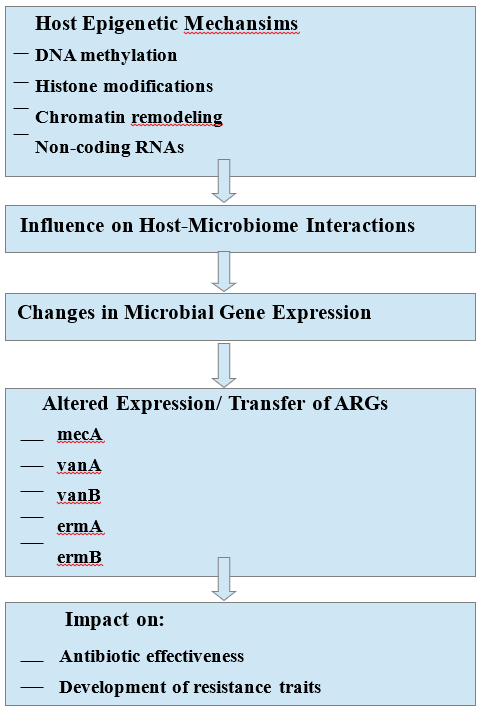
Legend 2. The Role of Host Epigenetics in the Expression of Antibiotic Resistance Genes (ARGs). The diagram suggests that host epigenetic mechanisms like DNA methylation, histone modification, or chromatin remodeling could influence the expression of microbial ARGs such as mecA, vanA, vanB, ermA, or ermB. This epigenetic regulation in the host may affect the microbiome, subsequently influencing the expression or transfer of these genes, which has implications for the effectiveness of antibiotics and the development of resistance traits.
To sum up, host epigenetic regulation is crucial in influencing the progression of S. aureus infections and its resistance to antibiotics. Epigenetic alterations in immune cells or epithelial defenses can diminish immune responses, facilitate bacterial colonization, and result in repeated infections. The microenviornments promote the survival of resistance strains, boost HGT, and facilitate the selection of resistance traits. Interactions between host and pathogen at the epigenetic level could be comprehended, potentially leading to innovative treatments that address both the pathogen and the host surroundings to prevent resistance and long-lasting infections.
Significance of the Study
While extensive knowledge exists regarding Staphylococcus aureus virulence mechanisms and antibiotic resistance, comparatively limited research has comprehensively examined the interplay between virulence determinants, bacterial regulatory systems, and host epigenetic modulation, and how these factors collectively facilitate immune evasion and antimicrobial resistance. In this review, we aim to integrate current insights from microbiology and immunology with knowledge of DNA methylation, histone modification, and non-coding RNAs, particularly in the context of the agr/sar regulatory axis and quorum-sensing–controlled virulence factors (e.g., Hla, PSMs, TSST-1) and toxin-mediated signaling components. We conceptualize S. aureus infection as an active molecular dialogue in which the pathogen induces real-time reprogramming of the host epigenome. Such reprogramming can drive immune phenotypic shifts toward either trained immunity or immune tolerance, promote bacterial persistence, and create optimal conditions for ARG expression, HGT, and MDR. This perspective proposes a novel feedback loop linking host epigenetic suppression and bacterial resistance and highlights potential therapeutic strategies beyond direct antimicrobial targeting—specifically, restoring or modulating host epigenetic states to potentiate existing treatments, limit persistence, and reduce the emergence of resistance. This conceptual framework occupies a distinct niche in the literature, positioning the host epigenome as both a target and an active participant in the evolutionary competition between S. aureus and the human immune system.
Future Directions
Exploring the epigenetic relationship between S. aureus and its human host offers promising avenues for fundamental research and clinical advancements. A particularly intriguing approach involves developing novel therapeutic strategies aimed at host epigenetic mechanisms alongside conventional antibiotics. By focusing on S. aureus-mediated immune suppression by regulating host epigenetics, these combination therapies can regain immune functionality, facilitate robust bacterial elimination, and prevent S. aureus from expressing ARGs. Incorporating epigenetic biomarkers into diagnostic tools might also improve clinicians' ability to predict infection outcomes, customize antimicrobial treatment, and take early action to prevent the development of S. aureus resistance. These approaches can be especially beneficial for high-risk groups—like the older population or individuals with weakened immune systems—by supporting immune health and reducing the burden of chronic infections and severity of disease.
One of the most promising yet least studied fields of research is the role of ARGs as potential modulators of the host's epigenetic response. While ARGs are typically studied concerning antimicrobial resistance (AMR), new evidence suggests that resistant S. aureus strains may exhibit greater virulence and persistence. Both of these traits can extend the duration of host-pathogen interactions, enabling the pathogen to epigenetically alter the host's immune cells [116,117]. Specific inquiries also exist regarding whether specific ARGs—or the mobile genetic elements that carry them—actively drive alterations in the epigenetic regulation of host genes [118,119]. In this regard, comparative epigenomic investigations of host cells infected with methicillin-resistant versus methicillin-sensitive S. aureus strains could yield an important understanding of how resistance features relate to immune modulation. Moreover, exploring whether secreted factors or small RNAs located proximal to ARGs can modify chromatin structure in immune cells may reveal novel strategies employed by S. aureus to evade immune detection. To further elaborate, notably fascinating is the conjecture that ARGs themselves—or stress signals induced by antibiotic pressure—could possibly initiate epigenetic reprogramming mechanisms of innate immune memory (trained immunity) or immunotolerance. This suggests a new hypothesis that ARG-carrying bacteria might not only protect against host defense systems but also alter them to improve microbial persistence. This research may identify previously unrecognized epigenetic susceptibilities that could serve as potential therapeutic targets, particularly in the context of persistent or antibiotic-resistant S. aureus infections.
Conclusion
S. aureus poses a formidable challenge in infectious disease management, functioning both as a common commensal and a potent pathogen. Its ability to transition between these states hinges on a finely tuned network of virulence regulators, including agr, sarA, and SigB. These systems enable S. aureus to swiftly adapt to host defenses, withstand environmental stressors, evade immune surveillance, and persist despite antibiotic pressure. Recent advances in epigenetics have unveiled an additional, highly intricate layer to this host–pathogen interplay. By manipulating host epigenetic machinery—through DNA methylation, histone modification, and ncRNA regulation—S. aureus can modulate immune responses, reprogram host cellular activities, and establish a microenvironment favorable for long-term survival. This emerging paradigm underscores the necessity of viewing S. aureus infections through an epigenetic lens, revealing previously unrecognized mechanisms of immune evasion and resistance. Such insights not only deepen our understanding of S. aureus pathogenesis but also open promising avenues for novel anti-virulence and host-directed therapeutics. Harnessing these epigenetic strategies could be transformative in combating persistent and drug-resistant S. aureus infections.
Conflict of Interest
The authors do not have anything to declare.
Funding
This research is supported by Bandhan, Kolkata, India.
Author Contribution
Conceptualization: S. K. C.; Formal analysis: S. K. C.; Original draft preparation: S. K. C.; Writing—review and editing: S. K. C. and D. C.; Supervision: S. K. C.; Project administration: S. K. C.; Funding acquisition: S. K. C.





p (1).png)




.png)




.png)
